Distributed Analysis with ROOT RDataFrame
The HL-LHC is foreseen to generate roughly thirty times more data than the LHC has produced so far. Given the available future budget estimations and expected technological evolution [1], software will play a crucial role to cover the performance gap and be able to process the foreseen data volumes.
In such a scenario, there will be a need for data analysis tools that can run both on a single local resource and on a set of distributed resources, while the programming model for either case stays the same and the complexity of distributed computation is hidden as much as possible. This article describes how ROOT addresses those challenges with the RDataFrame API for analysis.
One programming model, many backends
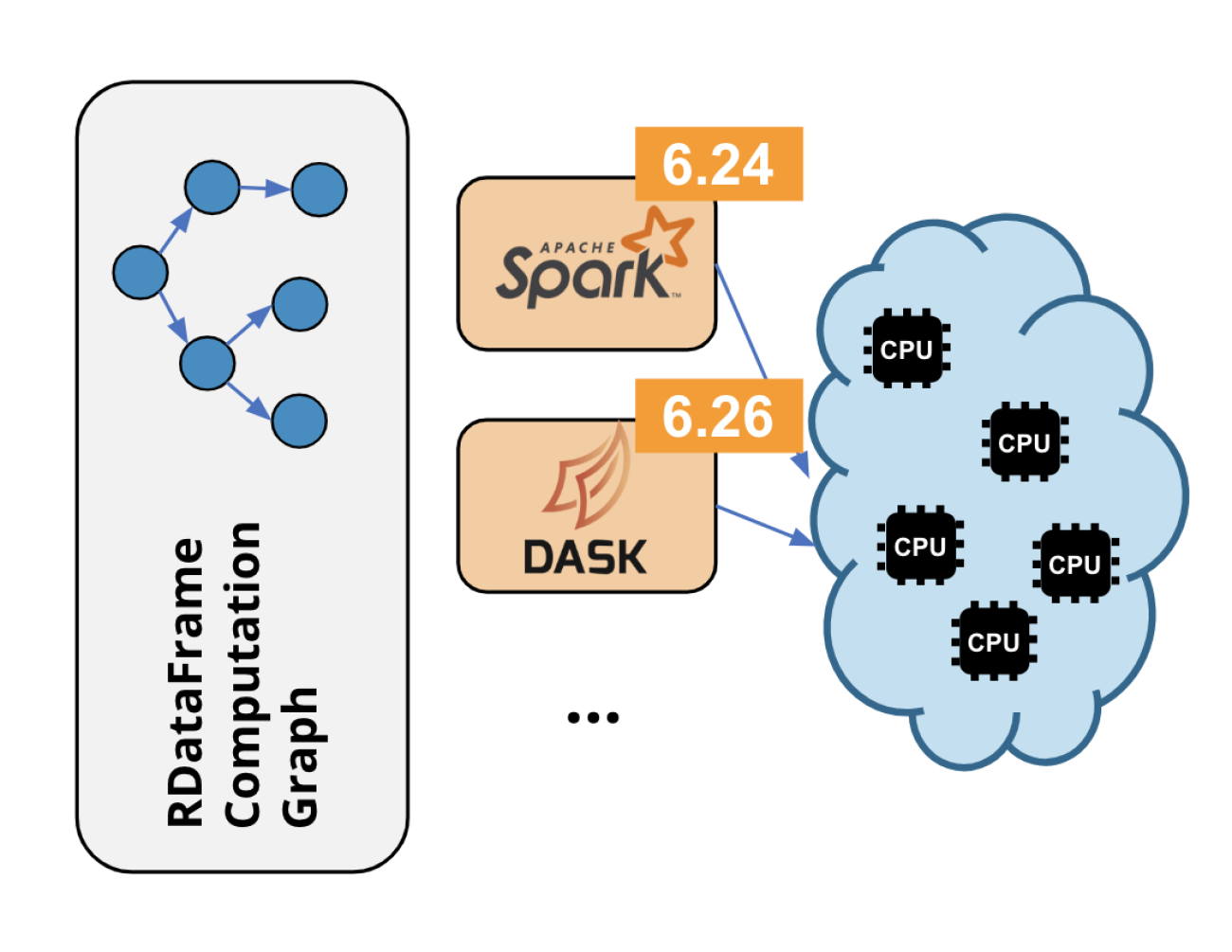
RDataFrame is ROOT’s high-level interface for data analysis since ROOT v6.14. By now many real world analyses use it, and on top of that we see lots of non-analysis usage in the wild. Parallelism has always been a staple of its design with support for executing the event loop on all cores of a machine thanks to implicit multi-threading. Since ROOT 6.24, this aspect of RDataFrame has been enhanced further with distributed computing capabilities, allowing users to run their analysis on multi-node computing clusters through widely used frameworks. Currently the package offers support for running the application on Apache Spark or Dask clusters, but the package design will make it possible to add many more backends in the future (e.g. AWS Lambda).
The main goal is to support distributed execution of any RDataFrame application. This has led to the creation of a Python package that connects the RDataFrame API (available in Python through PyROOT, ROOT’s Python bindings) and the APIs of distributed computing frameworks, which are offered in Python in the vast majority of cases. Another key goal is to offer a variety of backends, to provide a solution to a variety of use cases. This is achieved through a modular implementation that defines a generic task (representing the RDataFrame computation graph) to be executed on data. The input dataset is logically split into many ranges of entries, which will be sent to the distributed nodes for processing. Each range will then be paired to the generic task and submitted to the computing framework via a specific backend implementation. An added benefit of using RDataFrame is that the distributed tasks run C++ computations. This is made possible by PyROOT and cling, ROOT’s C++ interpreter.
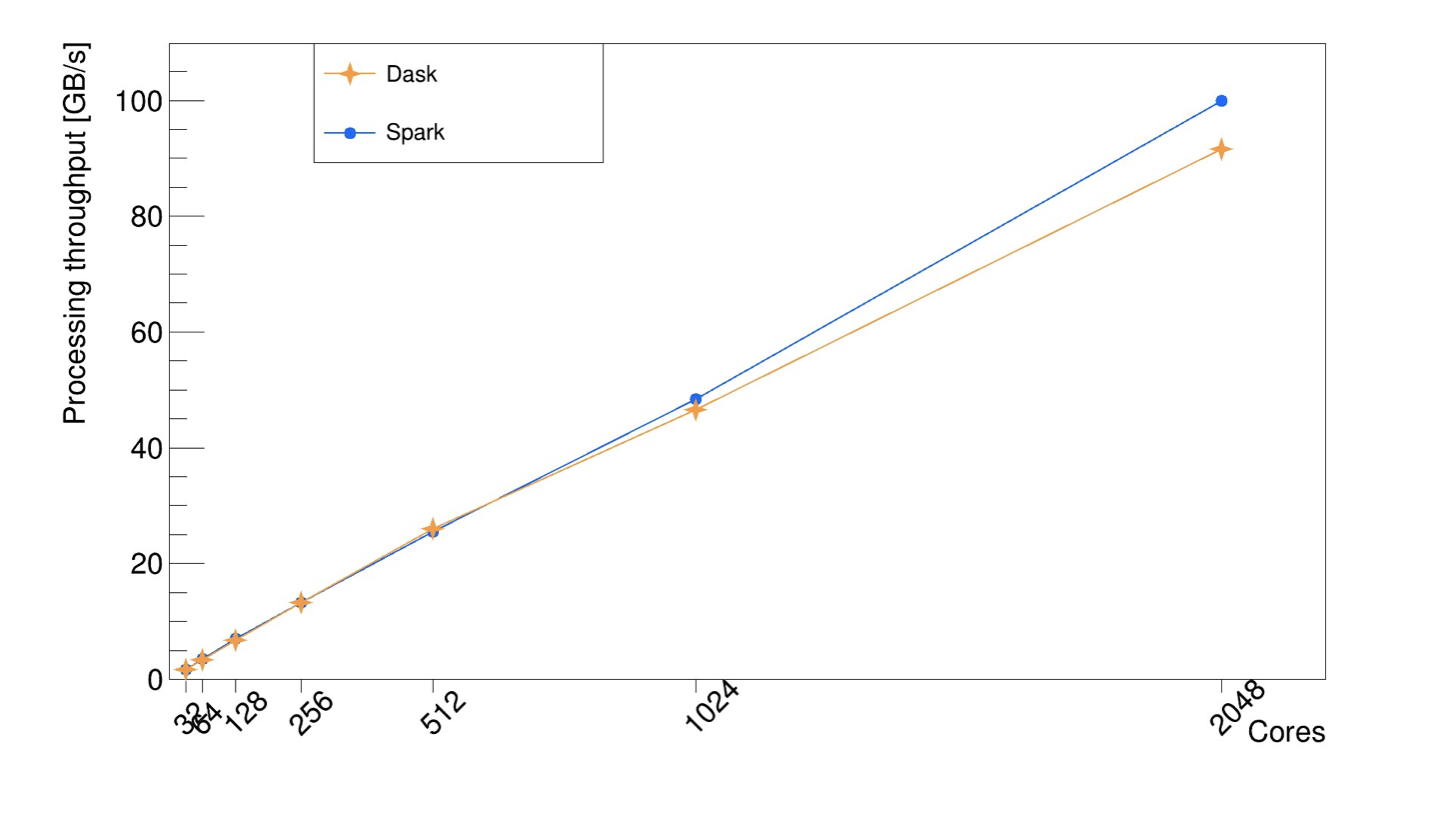
Excellent scaling is paramount for this distributed RDataFrame implementation, to ensure one can run the RDataFrame computation graph efficiently across multiple computing nodes and different backend implementations. This has been shown since the first stages of the development of this package with a real use case analysis running on a Spark cluster [2]. More recently, a benchmark based on CERN open data has shown promising scaling performance beyond 1000 cores with both Spark and Dask, as it can be seen in the plot below. The measures were taken in an HPC cluster.
How does a distributed analysis with RDataFrame actually look in code? Below is an example of analysis that uses the Spark backend:

In the example above, the only difference with respect to a local RDataFrame analysis is the usage of a Spark-specific RDataFrame, but the analysis itself – i.e. the operations on the input dataset – does not require any modification.
If instead of Spark we were to use Dask, and in particular we wanted to offload our computations to an existing Dask cluster, here is an example of how we would proceed:
from a dask.distributed import Client:
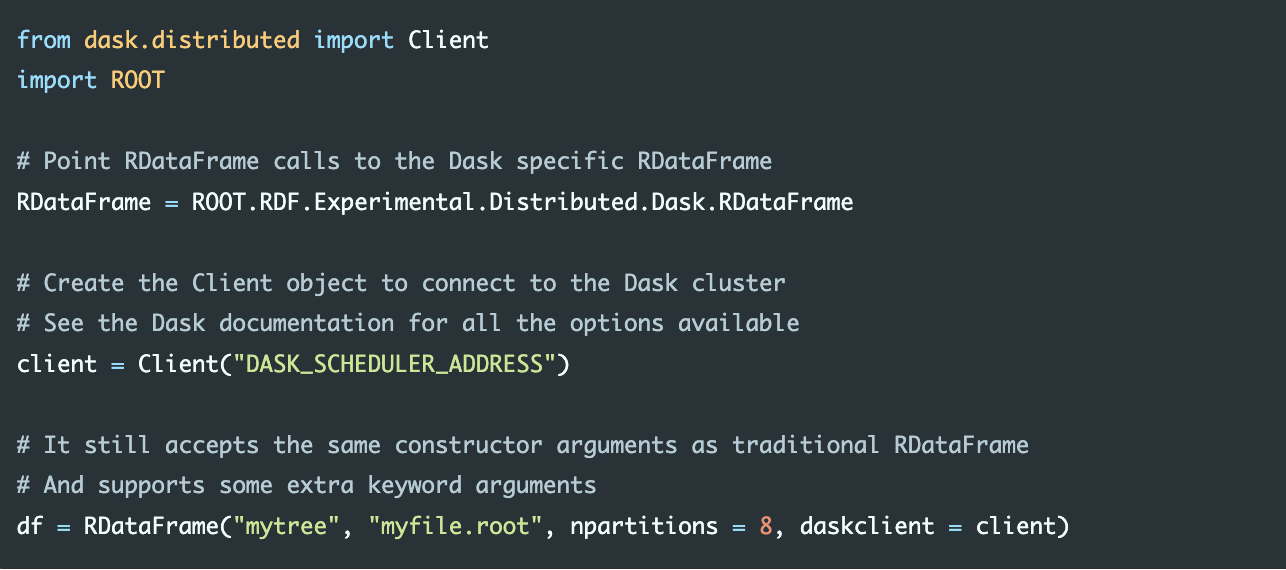
In this example the npartitions parameter tells the RDataFrame into how many ranges of entries the input dataset should be split. Each range will then correspond to a task on a node of the cluster. The daskclient parameter receives the object needed to connect to the Dask scheduler. All the options are available in the Dask documentation. The equivalent object for the Spark framework is called SparkContext and in general every backend will have its own way to connect to a cluster of nodes. Once the correct RDataFrame object has been created, there is no need to modify any other part of the program.
Conclusions
Distributed RDataFrame enables large-scale interactive data analysis with ROOT. This Python layer on top of RDataFrame allows to steer C++ computations on a set of computing nodes, returning the final result directly to the user, so that the entire analysis can be run within the same application. It supports all the basic features of local RDataFrame, such as filtering events, adding new columns or snapshotting datasets, and it is being extended to implement the distributed version of more advanced features such as systematic variations.
Distributed RDatraFrame is available as an experimental feature since ROOT 6.24 with the support for running on a Spark cluster, with the Dask backend introduced in 6.26 and more backends in the works. Try it with these two tutorials, one for Spark and one for Dask, and learn more about it in the respective RDataFrame documentation section.
References
[1] E. Elsen, “A Roadmap for HEP Software and Computing R&D for the 2020s,” Comput Softw Big Sci, vol. 16, no. 3, 2019.
[2] Valentina Avati et al. “Declarative Big Data Analysis for High-Energy Physics: TOTEM Use Case”. In:Euro-Par 2019: Parallel Processing (2019),pp. 241–255. https://doi.org/10.1007/978-3-030-29400-7_18
