RNTuple: The New Columnar Storage for HEP Data
R&D activities are vital for the ROOT project. A few years ago, we started investigating the evolution of TTree, ROOT’s columnar format with which more than two exabytes of data were written at the LHC: experiments write data, lots of it, to then store them on media such as disk or tape. A storage challenge is ahead of us in the HL-LHC era, during which experiments will acquire about ten times more data than the data collected by the end of Run 3.
It is indeed also to address that challenge that we started working on RNTuple. You may have heard of it, from CHEP’24, from other talks, or from our previous ROOT blog post, or maybe even already tried it out yourself. In case you haven’t: RNTuple is ROOT’s new I/O system for event data. Think of it as TTree, but more compact, faster, modern and more robust. With respect to TTree, we routinely see file size reductions between 10%-50%, multiple factors faster read throughput, and much better write performance and multicore scalability. RNTuple can fully harness the performance of modern NVMe drives and object stores, and it comes with a modern, safe, and feature rich API.
First Release of the RNTuple On-Disk Format
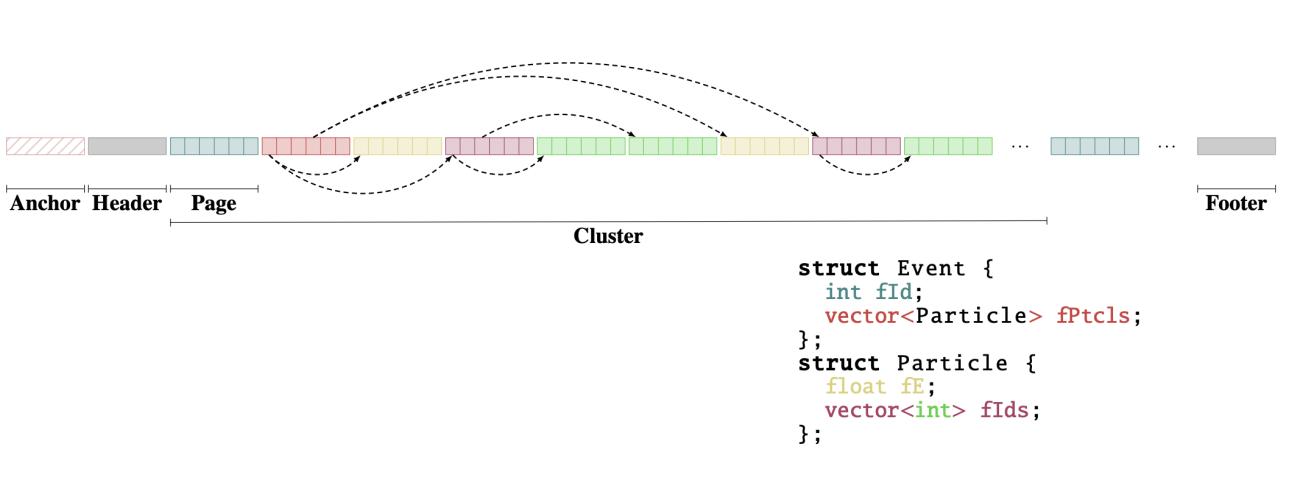
After 6 years of R&D, RNTuple reached an important milestone: we released the first official version of the on-disk binary format! The first ROOT version where this format is honoured is ROOT 6.34, released in November 2024. What does it mean for you, in a nutshell? It means that all future ROOT versions will be able to read back such objects. That may sound trivial, yet it has significant implications behind the scenes. The on-disk format, to recap, defines the layout of the bits on storage media that hold a certain data set. If we want to store, say, a collection of particles for every event, the on-disk format decides things like the endianness of the numerical values used to describe the properties of these particles, whether we store all particles consecutively or group the values of their properties (such as transverse momentum) together [1], how we remember the in-memory layout of the C++ class representing a particle, the compression block size, where we put data checksums, and so forth. Once the format is fixed and applications write (large amounts of) data, there are severe restrictions on what can still be practically changed.
Why then, you may ask, did we change at all from the TTree format? It was working very well after all, right? The reason is that some of RNTuple’s biggest advantages are intimately linked to the on-disk representation. Perhaps most importantly: the savings in data volume. For instance, for both CMS NanoAODs and ATLAS PHYSLITE files, compared to TTree we can store the same content in substantially fewer bytes:
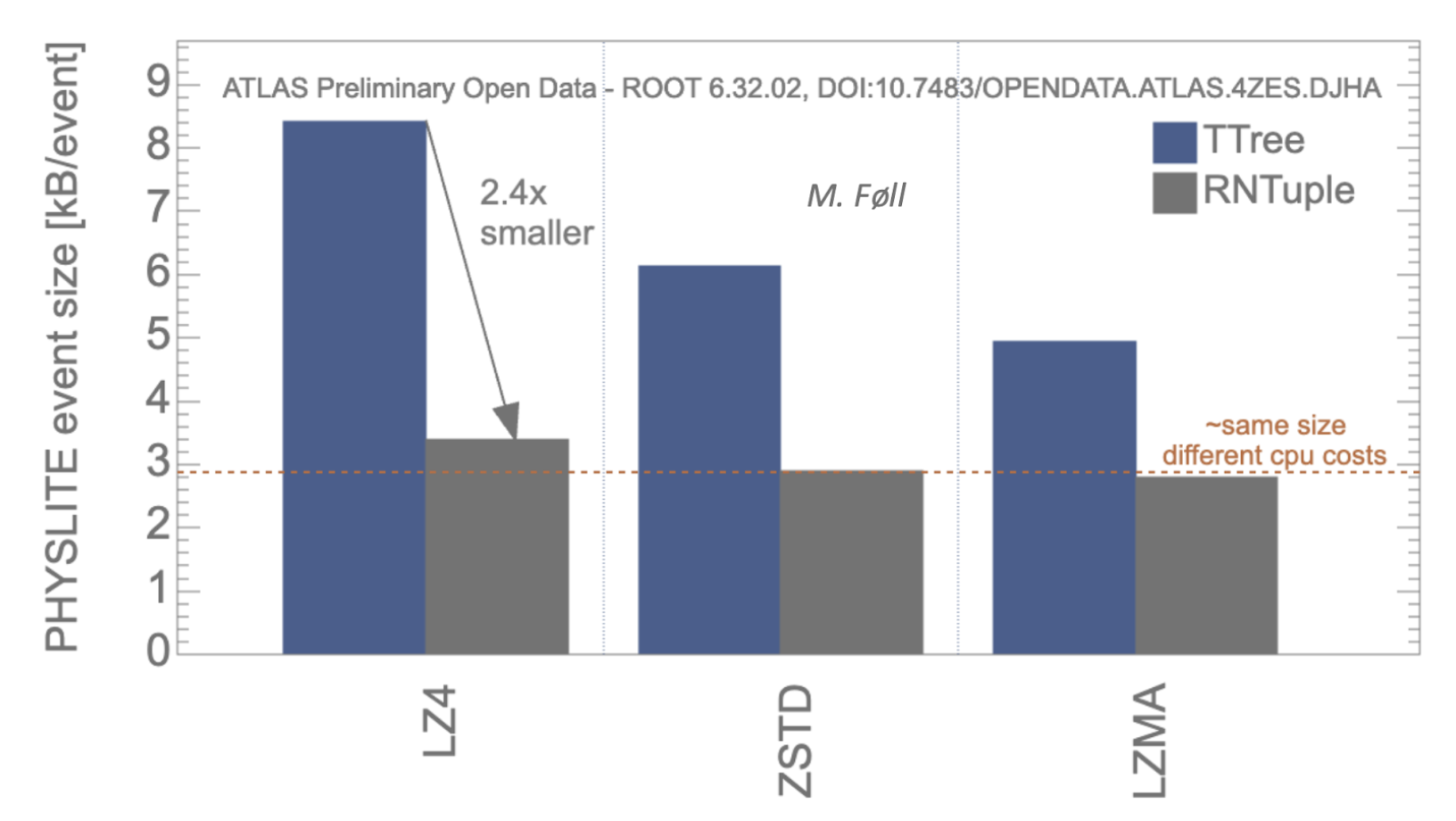
Figure 1: A plot of the size per event of ATLAS analysis data model, PHYSLITE. Three different compression algorithms are investigated, where LZMA is the most CPU consuming and LZ4 the least CPU consuming. Besides the reduction of the size per event when moving from TTree, blue tall columns, and RNTuple, gray small columns, it’s worth noting that, thanks to the preconditioning of the data performed by RNTuple, approximately the same size per event is obtained irrespective of the CPU invested in the compression: smaller event sizes with less computing power used!
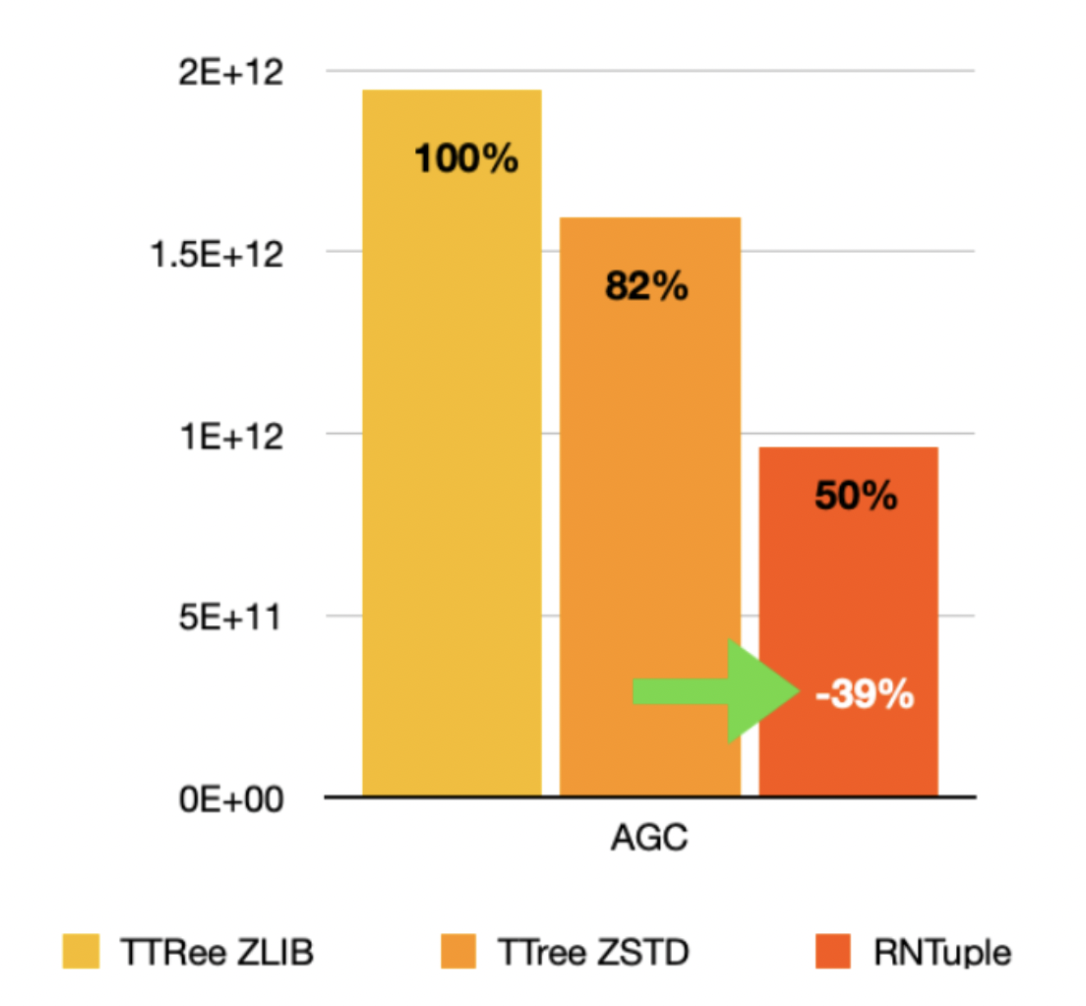
Figure 2: Total size (in bytes) of the Analysis Grand Challenge dataset, which is in CMS NanoAOD format. The first two columns represent the size when using TTree with two different compression algorithms. The second and the third, the size of the dataset compressed with the same compression algorithm, ZSTD, but with TTree and RNTuple: the new columnar dataset software technology results in a 40% reduction of the size!
This, and other reasons related to the robustness and the use of modern storage systems, convinced us to introduce a new on-disk format after more than 25 years of TTree. With RNTuple, we aim at a new, stable format for the exabytes of data to be expected during the lifetime of upcoming experiments (e.g., the experiments at the HL-LHC, EIC, DUNE, and beyond).[2]
Obviously, designing for such long time spans requires adding the provisions to adapt to future, not-yet predictable, environments. For RNTuple, we put mechanisms in place for forward- and backward-compatibility to enable new software versions to read old data and old software versions to (partially) read new data. For your own data models, you may know such mechanisms under the name “schema evolution”. For the RNTuple format itself, similar techniques are implemented in order to add, for instance, optional information such as metadata in the future.
Some other key features of the new format are
- a formal specification, allowing third parties to easily write data readers and writers without having to reverse engineer the code;
- strict use of 64 bit checksums both on data and metadata, ensuring data integrity;
- forward-looking limits that prepare us, e.g., for storing hundreds of thousands of columns in files of hundreds of terabytes each;
- preparation for native use of object stores such as S3 or DAOS;
- support for new C++ types, such as std::variant, std::optional, and the upcoming half precision floating point std::float16_t.
Now, you may wonder how to actually read and write RNTuples. To do so, RNTuple comes with a new set of APIs, too! Modern, safe, and easy to use correctly (so we hope!). An external API review, a first for the ROOT team, conducted by the High Energy Physics Center for Computational Excellence just concluded. We expect to see a first set of production APIs in ROOT 6.36.00, scheduled for the second quarter of 2025. Meanwhile, you can try out the current state and tell us what you think! The ATLAS, CMS, and LHCb experiments already include initial support for the API, so you may get your next RNTuple from your experiment. If you have analysis code in RDataFrame: job done. RDataFrame transparently processes TTree and RNTuple datasets interchangeably - by choosing to write your analysis with RDataFrame, you are guaranteed a seamless transition to RNTuple. We just require you to change the name of the ROOT files to the ones containing the columnar datasets in the RNTuple format!
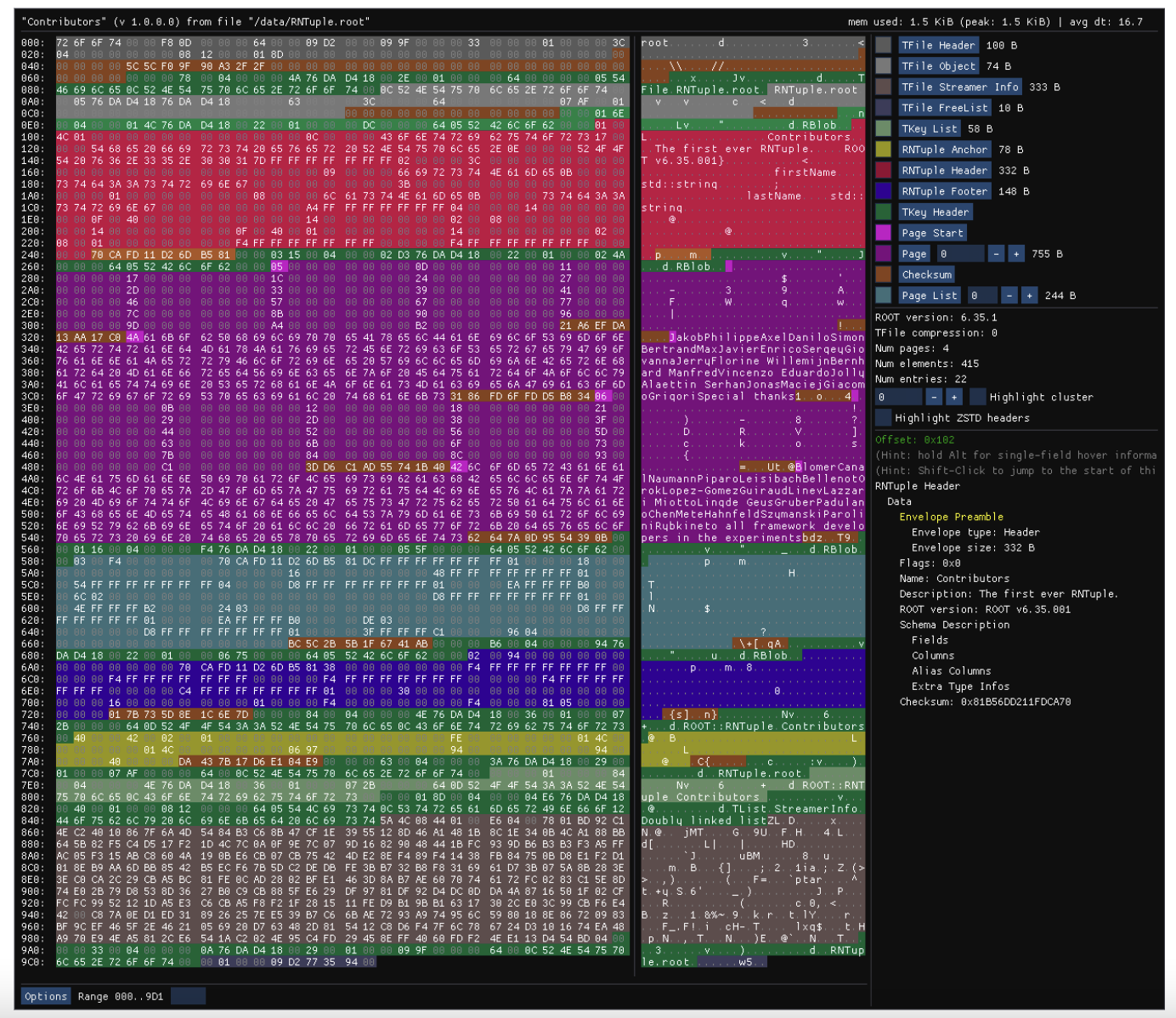
To close this article, I’d like to show you the first RNTuple ever written with the official specification. What you see is the hex dump of https://root.cern/files/RNTuple.root, visualized with the RNTuple viewer. You’d like to know more? Open the file in a ROOT release of the 6.34 cycle, for instance with the new RBrowser!
Notes
[1] ROOT does and has always done the latter. It is sometimes called columnar storage and it allows, e.g., to efficiently plot only pt without having to read charge.
[2] TTree, of course, will remain supported in ROOT! So you don’t need to worry about existing ROOT files.
